Bootstrap Current Self-Consistency
This tutorial demonstrates how to optimize a quasi-symmetric equilibrium to have a self-consistent bootstrap current profile. This is performed by minimizing the difference between the toroidal currents \(\langle J \cdot B \rangle\) computed from the MHD equilibrium and from the Redl formula. The Redl formula is only valid in the limit of perfect quasi-symmetry, so this procedure will not work for other configurations that are not quasi-symmetric.
There are two methods that can be used, and both will be shown:
Optimize the current profile for self-consistency
Iteratively solve the equilibrium with new current profiles
These methods should be equivalent, although one might be faster than the other depending on the particular problem.
If you have access to a GPU, uncomment the following two lines:
[12]:
# from desc import set_device
# set_device("gpu")
[13]:
import sys
import os
sys.path.insert(0, os.path.abspath("."))
sys.path.append(os.path.abspath("../../../"))
[14]:
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams["font.size"] = 14
from desc.compat import rescale
from desc.equilibrium import EquilibriaFamily
from desc.examples import get
from desc.grid import LinearGrid
from desc.objectives import (
BootstrapRedlConsistency,
FixAtomicNumber,
FixBoundaryR,
FixBoundaryZ,
FixCurrent,
FixElectronDensity,
FixElectronTemperature,
FixIonTemperature,
FixPsi,
ForceBalance,
ObjectiveFunction,
)
from desc.plotting import plot_1d
from desc.profiles import PowerSeriesProfile
As an example, we will reproduce the QA results from Landreman et al. (2022).
We will start with the “precise QA” example equilibrium, scaled to the ARIES-CS reactor size.
[15]:
eq0 = get("precise_QA")
eq0 = rescale(eq0, L=("R0", 10), B=("B0", 5.86))
This equilibrium has the vacuum profiles \(p = 0 ~\text{Pa}\) and \(\frac{2\pi}{\mu_0} I = 0 ~\text{A}\). Calculating the bootstrap current requires knowledge of the temperature and density profiles for each species in the plasma. We replace the pressure profile with the following kinetic profiles corresponding to \(\langle\beta\rangle=2.5\%\):
\(n_e = n_i = 2.38\times10^{20} (1 - \rho^{10}) ~\text{m}^{-3}\)
\(T_e = T_i = 9.45\times10^{3} (1 - \rho^{2}) ~\text{eV}\)
The temperature profiles must be given for both ions and electrons, but only the electron density profile is specified. The ion density profile is given by the effective atomic number \(Z_{eff}\) as \(n_i = n_e / Z_{eff}\). The plasma pressure will then be computed as
\(p = e (n_e T_e + n_i T_i)\).
[16]:
eq0.pressure = None # must remove the pressure profile before setting kinetic profiles
eq0.atomic_number = PowerSeriesProfile([1])
eq0.electron_density = PowerSeriesProfile(params=[1, -1], modes=[0, 10]) * 2.38e20
eq0.electron_temperature = PowerSeriesProfile(params=[1, -1], modes=[0, 2]) * 9.45e3
eq0.ion_temperature = PowerSeriesProfile(params=[1, -1], modes=[0, 2]) * 9.45e3
# the existing current profile is the vacuum case eq0.current = PowerSeriesProfile([0])
We need to re-solve the equilibrium force balance with the new profiles.
[17]:
eq0, _ = eq0.solve(objective="force", optimizer="lsq-exact", verbose=3)
Building objective: force
Precomputing transforms
Timer: Precomputing transforms = 867 ms
Timer: Objective build = 1.58 sec
Timer: Linear constraint projection build = 8.56 sec
Compiling objective function and derivatives: ['force']
Timer: Objective compilation time = 9.30 sec
Timer: Jacobian compilation time = 12.3 sec
Timer: Total compilation time = 21.6 sec
Number of parameters: 856
Number of objectives: 5346
Starting optimization
Using method: lsq-exact
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 1.506e-02 1.077e+01
1 4 2.845e-04 1.478e-02 9.222e-02 2.691e+00
2 5 1.830e-04 1.014e-04 1.397e-01 1.249e+00
3 6 1.089e-04 7.414e-05 1.213e-01 7.621e-01
4 7 3.200e-05 7.690e-05 9.716e-02 6.395e-01
5 8 1.869e-05 1.331e-05 9.350e-02 2.676e-01
6 10 1.651e-06 1.704e-05 2.585e-02 7.973e-02
7 12 6.750e-07 9.757e-07 1.704e-02 6.897e-02
8 14 4.748e-07 2.002e-07 7.785e-03 1.575e-02
9 15 4.482e-07 2.658e-08 1.338e-02 9.857e-03
10 16 4.128e-07 3.545e-08 1.298e-02 2.335e-03
11 18 3.932e-07 1.959e-08 6.564e-03 5.648e-04
12 19 3.792e-07 1.398e-08 1.314e-02 1.775e-03
13 20 3.616e-07 1.763e-08 1.309e-02 1.628e-03
14 21 3.464e-07 1.518e-08 1.294e-02 1.850e-03
15 22 3.335e-07 1.289e-08 1.267e-02 2.304e-03
16 23 3.226e-07 1.093e-08 1.239e-02 2.642e-03
17 24 3.137e-07 8.889e-09 1.217e-02 2.861e-03
18 25 3.081e-07 5.589e-09 1.204e-02 3.115e-03
19 27 2.977e-07 1.040e-08 3.093e-03 3.110e-04
20 28 2.960e-07 1.678e-09 5.981e-03 1.224e-03
Optimization terminated successfully.
`ftol` condition satisfied.
Current function value: 2.960e-07
Total delta_x: 5.126e-01
Iterations: 20
Function evaluations: 28
Jacobian evaluations: 21
Timer: Solution time = 1.03 min
Timer: Avg time per step = 2.95 sec
Start of solver
Total (sum of squares): 1.506e-02,
Maximum absolute Force error: 6.502e+07 (N)
Minimum absolute Force error: 2.968e-01 (N)
Average absolute Force error: 5.342e+06 (N)
Maximum absolute Force error: 2.008e-02 (normalized)
Minimum absolute Force error: 9.164e-11 (normalized)
Average absolute Force error: 1.650e-03 (normalized)
R boundary error: 0.000e+00 (m)
Z boundary error: 0.000e+00 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
End of solver
Total (sum of squares): 2.960e-07,
Maximum absolute Force error: 9.149e+05 (N)
Minimum absolute Force error: 5.582e+00 (N)
Average absolute Force error: 3.383e+04 (N)
Maximum absolute Force error: 2.825e-04 (normalized)
Minimum absolute Force error: 1.724e-09 (normalized)
Average absolute Force error: 1.045e-05 (normalized)
R boundary error: 6.836e-17 (m)
Z boundary error: 2.883e-17 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
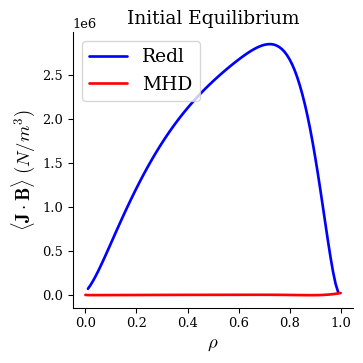
Now we have our initial equilibrium, which does not have a self-consistent bootstrap current:
[18]:
fig, ax = plot_1d(eq0, "<J*B> Redl", linecolor="b", lw=2, label="Redl")
fig, ax = plot_1d(eq0, "<J*B>", linecolor="r", lw=2, label="MHD", ax=ax)
ax.legend(loc="best")
ax.set_title("Initial Equilibrium")
[18]:
Text(0.5, 1.0, 'Initial Equilibrium')
/home/dudt/anaconda3/envs/desc/lib/python3.10/site-packages/IPython/core/pylabtools.py:152: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.canvas.print_figure(bytes_io, **kw)
We need to create a grid on which to evaluate the boootstrap current self-consistency. The bootstrap current is a radial profile, but the grid must have finite poloidal and toroidal resolution to accurately compute flux surface quantities. The Redl formula is undefined where the kinetic profiles vanish, so in our example we do not include points at \(\rho=0\) or \(\rho=1\).
[19]:
grid = LinearGrid(
M=eq0.M_grid,
N=eq0.N_grid,
NFP=eq0.NFP,
sym=eq0.sym,
rho=np.linspace(1 / eq0.L_grid, 1, eq0.L_grid) - 1 / (2 * eq0.L_grid),
)
Our current profile will be represented as a power series of the form:
\(I = c_0 + c_1 \rho + c_2 \rho^2 + \mathcal{O}(\rho^3)\)
Physically, the current should vanish on the magnetic axis so \(c_0 = 0\). And in order for the MHD equilibrium to be analytic, it should scale as \(\mathcal{O}(\rho^2)\) near the magnetic axis so \(c_1 = 0\) also. However, the Redl bootstrap current formula scales as \(\mathcal{O}(\sqrt{\rho})\) near the magnetic axis. This is incorrect, because the drift-kinetic equation from the Redl formula does not account for finite orbit width effects that become important near the axis.
Typically, we use even power series with sym=True for all equilibrium profiles to give the desired analycity conditions. For bootstrap current optimizations, it is recommended to use the full power series with sym=False while also enforcing \(c_0 = c_1 = 0\). This prevents getting good self-consistency near the magnetic axis, but allows for good agreement throughout the rest of the plasma volume and results in high quality equilibria overall.
[20]:
eq0.current = PowerSeriesProfile(np.zeros((eq0.L + 1,)), sym=False)
1. Optimization
In this method, we will optimize the current profile to minimize the self-consistency errors evaluated by the BootstrapRedlConsistency objective. This objective requires the helicity, which for QA is \((M, N) = (1, 0)\).
In this example we will only optimize the current profile, so all other profiles and the plasma boundary are constrained to be fixed. It is recommended to use a very small value for gtol when optimizing the bootstrap current.
[29]:
eq1 = eq0.copy()
[30]:
objective = ObjectiveFunction(
BootstrapRedlConsistency(eq=eq1, grid=grid, helicity=(1, 0)),
)
constraints = (
FixAtomicNumber(eq=eq1),
FixBoundaryR(eq=eq1),
FixBoundaryZ(eq=eq1),
FixCurrent(eq=eq1, indices=[0, 1]), # fix c_0=c_1=0 current profile coefficients
FixElectronDensity(eq=eq1),
FixElectronTemperature(eq=eq1),
FixIonTemperature(eq=eq1),
FixPsi(eq=eq1),
ForceBalance(eq=eq1),
)
eq1, _ = eq1.optimize(
objective=objective,
constraints=constraints,
optimizer="proximal-lsq-exact",
maxiter=4,
gtol=1e-16, # it is recommended to use a very small gtol
verbose=3,
)
Building objective: Bootstrap current self-consistency (Redl)
Precomputing transforms
Timer: Precomputing transforms = 370 ms
Timer: Objective build = 1.64 sec
Building objective: force
Precomputing transforms
Timer: Precomputing transforms = 288 ms
Timer: Objective build = 843 ms
Timer: Proximal projection build = 10.1 sec
Timer: Linear constraint projection build = 8.22 sec
Compiling objective function and derivatives: ['Bootstrap current self-consistency (Redl)']
Timer: Objective compilation time = 7.42 sec
Timer: Jacobian compilation time = 55.4 sec
Timer: Total compilation time = 1.04 min
Compiling objective function and derivatives: ['force']
Timer: Objective compilation time = 3.03 sec
Timer: Jacobian compilation time = 10.7 sec
Timer: Total compilation time = 13.8 sec
Number of parameters: 7
Number of objectives: 16
Starting optimization
Using method: proximal-lsq-exact
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 2.258e-02 1.935e-08
1 2 9.430e-05 2.249e-02 1.237e+07 1.029e-09
2 3 2.292e-06 9.201e-05 2.480e+07 4.206e-12
3 4 1.750e-06 5.424e-07 4.868e+07 1.039e-11
4 5 1.477e-06 2.729e-07 6.573e+07 8.437e-12
Warning: Maximum number of iterations has been exceeded.
Current function value: 1.477e-06
Total delta_x: 1.391e+08
Iterations: 4
Function evaluations: 5
Jacobian evaluations: 5
Timer: Solution time = 2.74 min
Timer: Avg time per step = 32.9 sec
Start of solver
Total (sum of squares): 2.258e-02,
Maximum absolute Bootstrap current self-consistency error: 2.840e+06 (T A m^-2)
Minimum absolute Bootstrap current self-consistency error: 1.677e+05 (T A m^-2)
Average absolute Bootstrap current self-consistency error: 1.787e+06 (T A m^-2)
Maximum absolute Bootstrap current self-consistency error: 3.027e-01 (normalized)
Minimum absolute Bootstrap current self-consistency error: 1.788e-02 (normalized)
Average absolute Bootstrap current self-consistency error: 1.905e-01 (normalized)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
R boundary error: 0.000e+00 (m)
Z boundary error: 0.000e+00 (m)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-Psi error: 0.000e+00 (Wb)
Maximum absolute Force error: 9.149e+05 (N)
Minimum absolute Force error: 5.582e+00 (N)
Average absolute Force error: 3.383e+04 (N)
Maximum absolute Force error: 2.857e-04 (normalized)
Minimum absolute Force error: 1.743e-09 (normalized)
Average absolute Force error: 1.056e-05 (normalized)
End of solver
Total (sum of squares): 1.477e-06,
Maximum absolute Bootstrap current self-consistency error: 3.941e+04 (T A m^-2)
Minimum absolute Bootstrap current self-consistency error: 3.852e+03 (T A m^-2)
Average absolute Bootstrap current self-consistency error: 1.294e+04 (T A m^-2)
Maximum absolute Bootstrap current self-consistency error: 4.200e-03 (normalized)
Minimum absolute Bootstrap current self-consistency error: 4.106e-04 (normalized)
Average absolute Bootstrap current self-consistency error: 1.379e-03 (normalized)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
R boundary error: 8.358e-17 (m)
Z boundary error: 6.953e-18 (m)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-Psi error: 0.000e+00 (Wb)
Maximum absolute Force error: 1.432e+06 (N)
Minimum absolute Force error: 1.249e+01 (N)
Average absolute Force error: 8.006e+04 (N)
Maximum absolute Force error: 4.472e-04 (normalized)
Minimum absolute Force error: 3.900e-09 (normalized)
Average absolute Force error: 2.500e-05 (normalized)
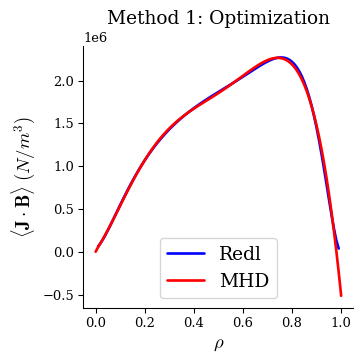
When plotting the bootstrap current profiles, we see the MHD equilibrium now has very good agreement with the Redl formula.
[31]:
fig, ax = plot_1d(eq1, "<J*B> Redl", linecolor="b", lw=2, label="Redl")
fig, ax = plot_1d(eq1, "<J*B>", linecolor="r", lw=2, label="MHD", ax=ax)
ax.legend(loc="best")
ax.set_title("Method 1: Optimization")
[31]:
Text(0.5, 1.0, 'Method 1: Optimization')
/home/dudt/anaconda3/envs/desc/lib/python3.10/site-packages/IPython/core/pylabtools.py:152: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.canvas.print_figure(bytes_io, **kw)
2. Iterative Solves
In this method, we iteratively solve the equilibrium with updated guesses for the current profile. The current profile is computed such that the parallel current is consistent with the Redl formula, according to Equation C3 in Landreman & Catto (2012). This is the same approach as STELLOPT VBOOT with SFINCS, and it usually converges in only a few iterations.
[24]:
eq2 = eq0.copy()
fam2 = EquilibriaFamily(eq2)
[25]:
niters = 3
for k in range(niters):
eq2 = eq2.copy()
# compute new guess for the current profile, consistent with Redl formula
data = eq2.compute("current Redl", grid)
current = grid.compress(data["current Redl"])
rho = grid.compress(data["rho"])
# fit the current profile to a power series, with c_0=c_1=0
XX = np.fliplr(np.vander(rho, eq2.L + 1)[:, :-2])
eq2.c_l = np.pad(np.linalg.lstsq(XX, current, rcond=None)[0], (2, 0))
# re-solve the equilibrium
eq2, _ = eq2.solve(objective="force", optimizer="lsq-exact", verbose=3)
fam2.append(eq2)
Building objective: force
Precomputing transforms
Timer: Precomputing transforms = 627 ms
Timer: Objective build = 1.65 sec
Timer: Linear constraint projection build = 7.71 sec
Compiling objective function and derivatives: ['force']
Timer: Objective compilation time = 2.80 sec
Timer: Jacobian compilation time = 9.89 sec
Timer: Total compilation time = 12.6 sec
Number of parameters: 856
Number of objectives: 5346
Starting optimization
Using method: lsq-exact
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 6.277e-03 8.832e+00
1 2 9.553e-04 5.322e-03 1.778e-01 2.882e+00
2 3 2.544e-04 7.009e-04 1.565e-01 8.031e-01
3 4 2.018e-04 5.260e-05 1.078e-01 2.517e+00
4 5 4.386e-06 1.975e-04 2.972e-02 1.524e-01
5 7 2.777e-06 1.609e-06 1.257e-02 2.818e-02
6 9 2.503e-06 2.742e-07 7.600e-03 1.122e-02
7 10 2.478e-06 2.540e-08 1.202e-02 3.106e-02
8 12 2.401e-06 7.660e-08 3.372e-03 3.923e-03
9 13 2.371e-06 3.045e-08 6.233e-03 2.433e-03
10 15 2.342e-06 2.853e-08 3.358e-03 1.067e-03
11 16 2.316e-06 2.603e-08 6.722e-03 7.377e-03
12 18 2.288e-06 2.836e-08 3.649e-03 2.782e-03
13 19 2.268e-06 1.946e-08 7.183e-03 9.851e-03
Optimization terminated successfully.
`ftol` condition satisfied.
Current function value: 2.268e-06
Total delta_x: 2.219e-01
Iterations: 13
Function evaluations: 19
Jacobian evaluations: 14
Timer: Solution time = 41.0 sec
Timer: Avg time per step = 2.93 sec
Start of solver
Total (sum of squares): 6.277e-03,
Maximum absolute Force error: 6.172e+07 (N)
Minimum absolute Force error: 3.756e+01 (N)
Average absolute Force error: 4.027e+06 (N)
Maximum absolute Force error: 1.706e-02 (normalized)
Minimum absolute Force error: 1.038e-08 (normalized)
Average absolute Force error: 1.113e-03 (normalized)
R boundary error: 0.000e+00 (m)
Z boundary error: 0.000e+00 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
End of solver
Total (sum of squares): 2.268e-06,
Maximum absolute Force error: 1.608e+06 (N)
Minimum absolute Force error: 9.463e+00 (N)
Average absolute Force error: 9.382e+04 (N)
Maximum absolute Force error: 4.443e-04 (normalized)
Minimum absolute Force error: 2.615e-09 (normalized)
Average absolute Force error: 2.593e-05 (normalized)
R boundary error: 2.867e-17 (m)
Z boundary error: 3.497e-18 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 9.313e-10 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
Building objective: force
Precomputing transforms
Timer: Precomputing transforms = 658 ms
Timer: Objective build = 1.56 sec
Timer: Linear constraint projection build = 7.61 sec
Compiling objective function and derivatives: ['force']
Timer: Objective compilation time = 4.68 sec
Timer: Jacobian compilation time = 9.66 sec
Timer: Total compilation time = 14.3 sec
Number of parameters: 856
Number of objectives: 5346
Starting optimization
Using method: lsq-exact
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 3.551e-04 2.865e+00
1 2 1.250e-05 3.426e-04 5.998e-02 8.708e-02
2 3 2.060e-06 1.044e-05 1.747e-02 2.468e-02
3 5 1.761e-06 2.993e-07 1.001e-02 1.671e-02
4 6 1.683e-06 7.778e-08 9.537e-03 5.701e-03
5 8 1.642e-06 4.113e-08 2.907e-03 1.141e-03
6 10 1.640e-06 1.511e-09 1.518e-03 2.400e-04
Optimization terminated successfully.
`ftol` condition satisfied.
Current function value: 1.640e-06
Total delta_x: 6.220e-02
Iterations: 6
Function evaluations: 10
Jacobian evaluations: 7
Timer: Solution time = 22.0 sec
Timer: Avg time per step = 3.14 sec
Start of solver
Total (sum of squares): 3.551e-04,
Maximum absolute Force error: 1.804e+07 (N)
Minimum absolute Force error: 4.405e+01 (N)
Average absolute Force error: 9.569e+05 (N)
Maximum absolute Force error: 5.075e-03 (normalized)
Minimum absolute Force error: 1.239e-08 (normalized)
Average absolute Force error: 2.691e-04 (normalized)
R boundary error: 0.000e+00 (m)
Z boundary error: 0.000e+00 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
End of solver
Total (sum of squares): 1.640e-06,
Maximum absolute Force error: 1.399e+06 (N)
Minimum absolute Force error: 9.554e+00 (N)
Average absolute Force error: 7.499e+04 (N)
Maximum absolute Force error: 3.934e-04 (normalized)
Minimum absolute Force error: 2.687e-09 (normalized)
Average absolute Force error: 2.109e-05 (normalized)
R boundary error: 2.776e-17 (m)
Z boundary error: 3.469e-18 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 1.547e-08 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
Building objective: force
Precomputing transforms
Timer: Precomputing transforms = 176 ms
Timer: Objective build = 502 ms
Timer: Linear constraint projection build = 3.70 sec
Compiling objective function and derivatives: ['force']
Timer: Objective compilation time = 2.77 sec
Timer: Jacobian compilation time = 8.01 sec
Timer: Total compilation time = 10.7 sec
Number of parameters: 856
Number of objectives: 5346
Starting optimization
Using method: lsq-exact
Iteration Total nfev Cost Cost reduction Step norm Optimality
0 1 1.201e-05 5.137e-01
1 2 1.968e-06 1.004e-05 2.063e-02 5.355e-02
2 3 1.683e-06 2.844e-07 1.181e-02 1.123e-02
3 5 1.630e-06 5.308e-08 4.613e-03 3.960e-03
4 6 1.627e-06 3.194e-09 3.133e-03 4.582e-04
Optimization terminated successfully.
`ftol` condition satisfied.
Current function value: 1.627e-06
Total delta_x: 1.640e-02
Iterations: 4
Function evaluations: 6
Jacobian evaluations: 5
Timer: Solution time = 22.0 sec
Timer: Avg time per step = 4.40 sec
Start of solver
Total (sum of squares): 1.201e-05,
Maximum absolute Force error: 2.969e+06 (N)
Minimum absolute Force error: 3.485e+01 (N)
Average absolute Force error: 1.823e+05 (N)
Maximum absolute Force error: 8.329e-04 (normalized)
Minimum absolute Force error: 9.777e-09 (normalized)
Average absolute Force error: 5.112e-05 (normalized)
R boundary error: 0.000e+00 (m)
Z boundary error: 0.000e+00 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 0.000e+00 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
End of solver
Total (sum of squares): 1.627e-06,
Maximum absolute Force error: 1.450e+06 (N)
Minimum absolute Force error: 3.668e+01 (N)
Average absolute Force error: 7.661e+04 (N)
Maximum absolute Force error: 4.067e-04 (normalized)
Minimum absolute Force error: 1.029e-08 (normalized)
Average absolute Force error: 2.149e-05 (normalized)
R boundary error: 2.776e-17 (m)
Z boundary error: 1.397e-20 (m)
Fixed-Psi error: 0.000e+00 (Wb)
Fixed-current profile error: 5.821e-11 (A)
Fixed-electron-density profile error: 0.000e+00 (m^-3)
Fixed-electron-temperature profile error: 0.000e+00 (eV)
Fixed-ion-temperature profile error: 0.000e+00 (eV)
Fixed-atomic-number profile error: 0.000e+00 (dimensionless)
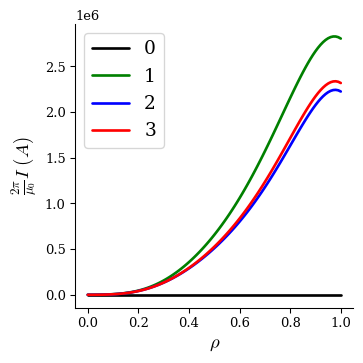
We can plot the current profile at each iteration to visualize how it changed:
[26]:
fig, ax = plot_1d(fam2[0], "current", linecolor="k", lw=2, label="0")
fig, ax = plot_1d(fam2[1], "current", linecolor="g", lw=2, label="1", ax=ax)
fig, ax = plot_1d(fam2[2], "current", linecolor="b", lw=2, label="2", ax=ax)
fig, ax = plot_1d(fam2[3], "current", linecolor="r", lw=2, label="3", ax=ax)
ax.legend(loc="best")
[26]:
<matplotlib.legend.Legend at 0x7fcfe256cee0>
/home/dudt/anaconda3/envs/desc/lib/python3.10/site-packages/IPython/core/pylabtools.py:152: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.canvas.print_figure(bytes_io, **kw)
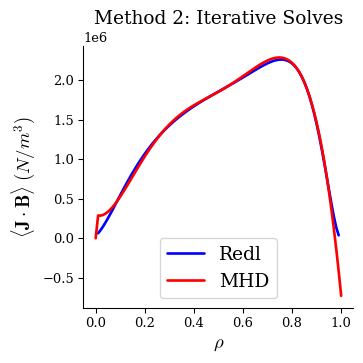
With this method the MHD equilibrium also has very good agreement with the Redl formula.
[27]:
fig, ax = plot_1d(eq2, "<J*B> Redl", linecolor="b", lw=2, label="Redl")
fig, ax = plot_1d(eq2, "<J*B>", linecolor="r", lw=2, label="MHD", ax=ax)
ax.legend(loc="best")
ax.set_title("Method 2: Iterative Solves")
[27]:
Text(0.5, 1.0, 'Method 2: Iterative Solves')
/home/dudt/anaconda3/envs/desc/lib/python3.10/site-packages/IPython/core/pylabtools.py:152: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.canvas.print_figure(bytes_io, **kw)
Comparison
Even though both methods give good self-consistency for the bootstrap current, they do result in slightly different coefficients for the current profile:
[32]:
print(eq1.c_l)
print(eq2.c_l)
[ 0.00000000e+00 0.00000000e+00 2.67019471e+03 3.97402482e+06
1.66002514e+07 -6.86550329e+07 1.01228131e+08 -6.28828607e+07
1.20716658e+07]
[ 0.00000000e+00 0.00000000e+00 4.36224944e+05 -1.62002569e+06
4.23552467e+07 -1.25717545e+08 1.66207488e+08 -9.88761406e+07
1.95335994e+07]
In this example, the first method of optimization gave better self-consistency but was noticeably slower than the second method of iterative solves.